How can AI promote the discovery of antibiotics?
Published:
How can AI promote the discovery of antibiotics?
The rapid development of antibiotic resistance in bacteria threatens public health globally more than ever. Nearly 5 million deaths yearly are related to drug-resistant bacteria, while 1.27 million of the annual deaths are directly linked to resistance to antimicrobials. To cope with the burden of antibacterial resistance, the use of antibiotics needs to be improved, or new antibacterial compounds should be discovered. Even though the process has its own economic and scientific challenges, researchers worldwide have been working diligently on discovering effective antimicrobial agents.
In this blog post, I would like to explore how machine-learning approaches can help discover drugs, such as antibiotics. In their article, Stokes et al. showed that a trained neural network model (DNNM) could track down a promising early-stage antibacterial molecule, halicin, by scanning numerous dug-like chemical libraries. I will review why and how they developed and applied a deep-learning algorithm in this manner.
Why is the help of AI needed for the discovery of antibiotics?
So, as it is widely known, antibiotics have played a crucial role in modern medicine since the discovery of penicillin by Alexander Fleming in 1928. Since this pioneering breakthrough, numerous antibiotics from divergent sources have been discovered and used clinically to fight against bacterial infections.
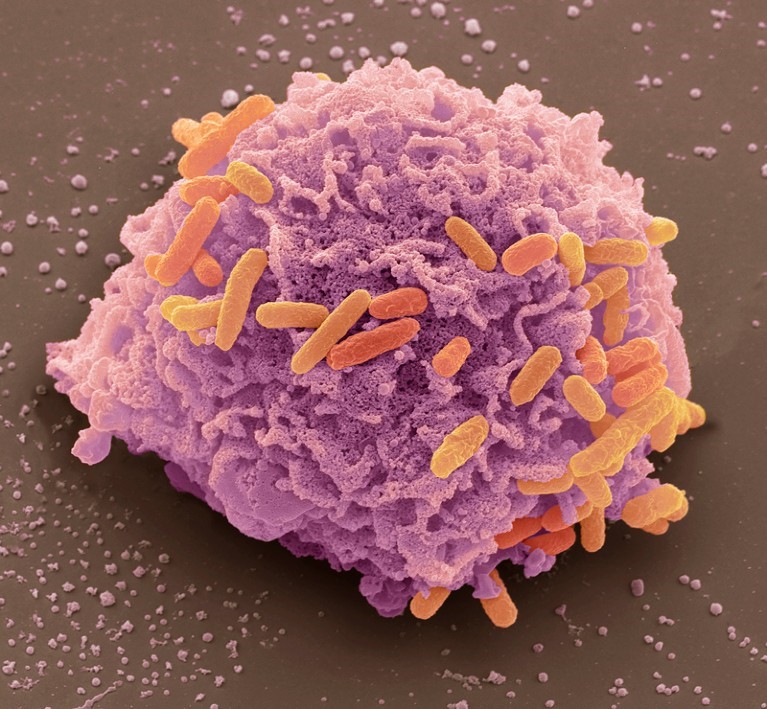 Figure-1 E.coli bacteria (orange) responsible for the resistant infections. Credit: Steve Gschmeissner
Figure-1 E.coli bacteria (orange) responsible for the resistant infections. Credit: Steve Gschmeissner
Over time, resistance to antibiotics has emerged as one of the biggest threats to global health by nature and accelerated by the misuse or overuse of drugs in the health and agriculture industry. Unfortunately, approximately 10 million deaths yearly from resistant infections are expected by 2050, alerting us to take urgent action for the identification of new and effective antibacterial drugs. Nonetheless, identifying and developing novel antibiotics are challenging for scientists and pharmaceutical companies, considering any new classes of antibacterial drugs have yet to be discovered in the last decades. Discovering and developing new antibiotics demands a vast investment of effort, time, and capital in screening natural or synthetic molecules, expensive and complex preclinical and clinical tests, approval as a drug, etc. Scientists have therefore sought innovative methods to enhance the rate of antibiotic discovery in a cost- and time-effective manner. One of the methods which can improve the discovery and development process is applying machine learning (ML) algorithms in combination with laboratory testing and bioinformatics analysis.
How does AI help to discover antibiotics?
 Figure-2 AI in medical sciences.
Figure-2 AI in medical sciences.
Nowadays, the application of AI and ML algorithms has started to be widely used in bio-related research and medical science with advents in high throughput screening of large data sets and libraries, identification of numerous structures or targets, classification of a vast number of molecules and making predictions more accurately by reducing time and money spent on trial and errors in the early stages.
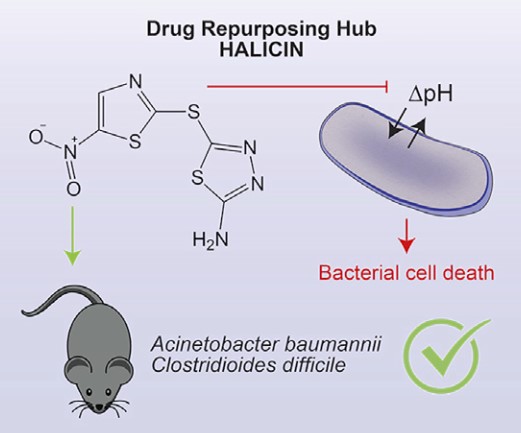
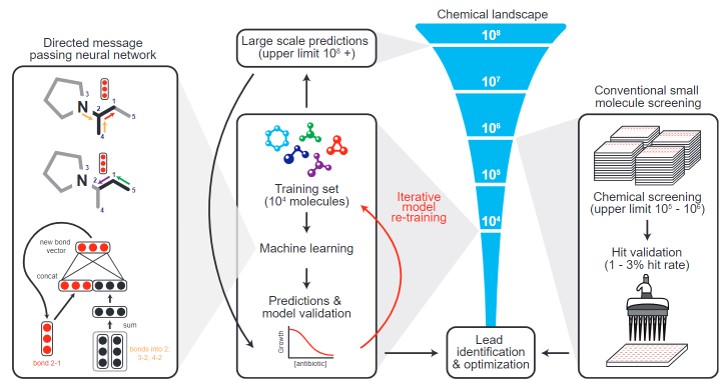 Figure-3 The graphical abstract of the paper.
Figure-3 The graphical abstract of the paper.
One of the latest applications of ML in this field came from a group of researchers at MIT. In their successive study, they developed a deep-learning approach and trained their neural network model to discover new antibacterial molecules. So, they first trained a deep neural network model by using a collection of 2335 molecules to predict bacterial growth inhibition. Next, they applied their model to various chemical libraries to identify the effective antibacterial substances and ranked the results according to the model predictions. Lastly, they selected the promising compounds based on the previously determined prediction score threshold, similarity in chemical structure to conventional antibiotics, and availability.
Now, you might ask the question of how an AI can screen chemical structures computationally in a systematic way. Herein lies the answer: molecular property prediction. I will not explain the details of molecular property prediction, but you find more about it here.
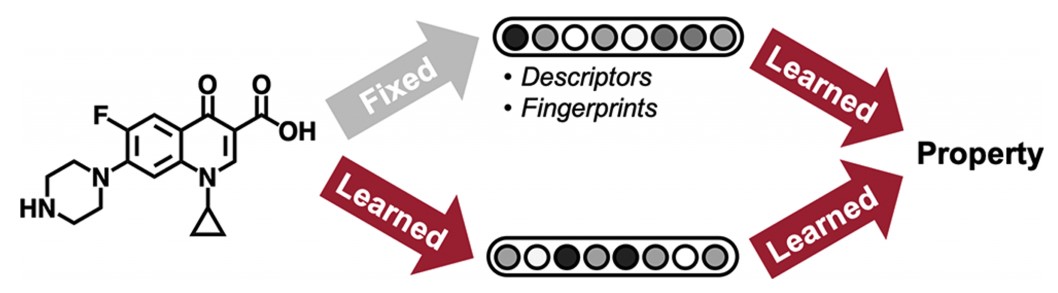
In their study, Stokes et al. used a previously developed graph convolutional direct message passing neural network model -Chemprop- to predict the molecular properties, where the molecules were represented in a graph structure (In the graph structure, atoms are depicted as nodes and bonds as edges. The computable properties of atoms and bonds were embedded in a feature vector for each atom and bond).
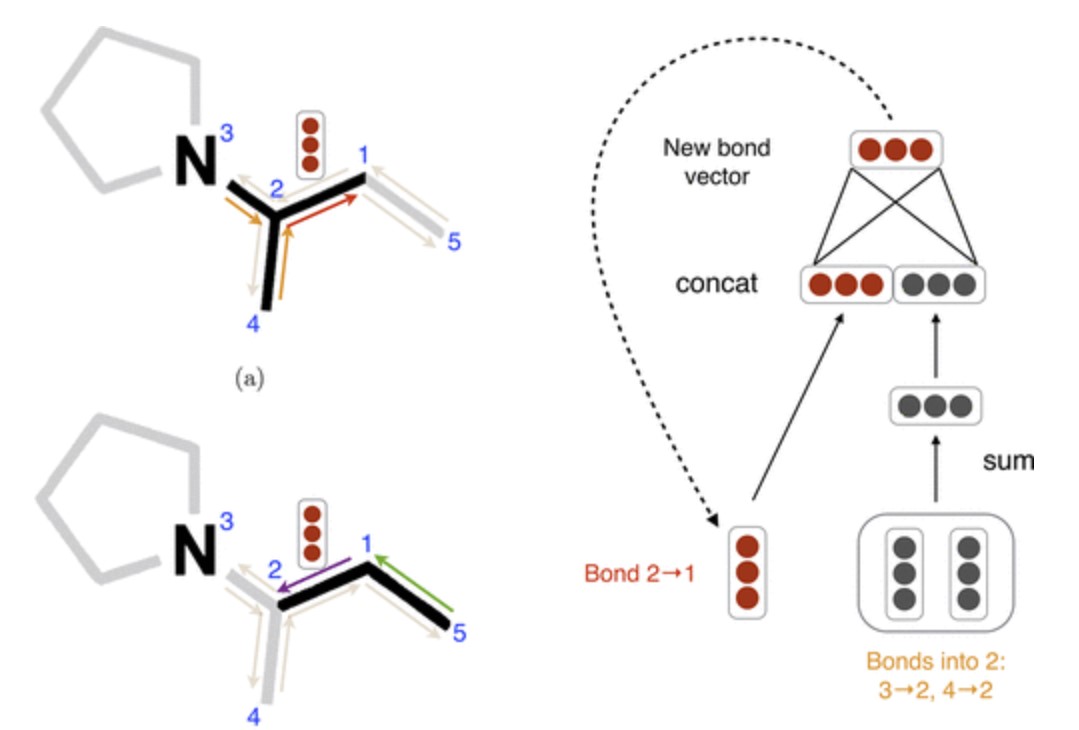 Figure-4 The bond level message passing in the model.
Figure-4 The bond level message passing in the model.
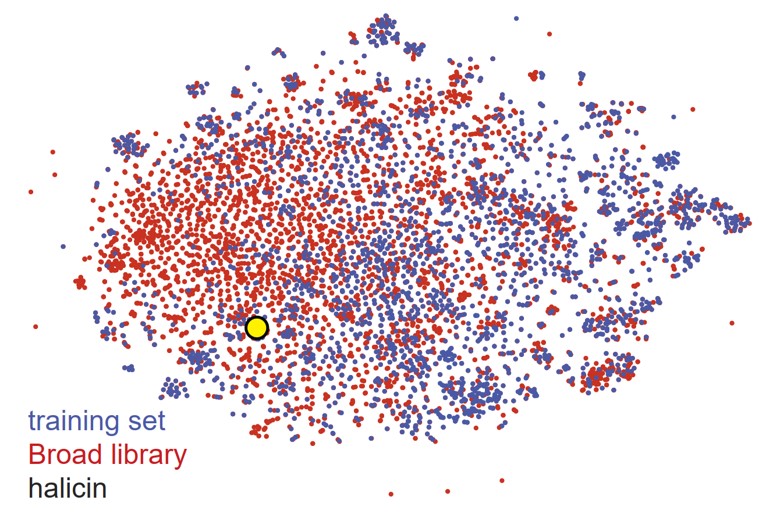
After training the model, they found a promising antibiotic molecule called halicin. It had a growth inhibitory activity on the bacteria E. coli BW25113, with low structural similarity to the current antibacterial drugs. They predicted toxicity among the molecules from the Drug Repurposing Hub consisting of 6111 molecules.
Then they tested the antibacterial activity of halicin over several pathogenic bacteria strains and showed that halicin has broad growth-inhibitory activity on antibiotic and drug-resistant pathogens. To prove that halicin could be a promising antibiotic candidate, they performed in-vivo experiments with murine models. As a result, they showed that halicin also has a broad-spectrum activity in mice.
At last, to move their work one step further, they screened an extensive chemical library: ZINC15 having approximately a hundred million molecules and with the retrained model, and discovered eight antibacterial molecules as potential antibiotics.
In a nutshell…
AI’s machine learning algorithms are impressive tools for the fast evaluation of biological properties of divergent molecules in chemical libraries with deep neural network-based methods. In their pioneering work, the researchers identified a promising antibacterial molecule, halicin, which is considered the antibiotic discovered with AI. Besides these, more importantly, they showed that AI is capable of screening a vast number of molecules and making predictions in a comparably shorter time, with less cost and effort than conventional methods.
References
[1] J. M. Stokes et al., “A Deep Learning Approach to Antibiotic Discovery,” Cell, vol. 180, no. 4, pp. 688-702.e13, Feb. 2020, doi: 10.1016/j.cell.2020.01.021.
[2] C. J. Murray et al., “Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis,” The Lancet, vol. 399, no. 10325, pp. 629–655, Feb. 2022, doi: 10.1016/S0140-6736(21)02724-0.
[3] L. L. Silver, “Challenges of antibacterial discovery,” Clin Microbiol Rev, vol. 24, no. 1, pp. 71–109, Jan. 2011, doi: 10.1128/CMR.00030-10.
[4] J. Vamathevan et al., “Applications of machine learning in drug discovery and development,” Nature Reviews Drug Discovery, vol. 18, no. 6. Nature Publishing Group, pp. 463–477, Jun. 01, 2019. doi: 10.1038/s41573-019-0024-5.
